The Rutgers Univeristy Micrboial Flow Sort Lab is particle analysis facility which utilizes flow cytometery and high speed cell sorting to assess a wide array of research questions from within the Rutgers scientific community and beyond. The Influx Mariner 209S Flow Cytometer uses the latest version BD FACS Sortware software for data acquisition. FlowJo [link] is the software used to read flow cytometry data which “facilitates complex data analysis in a graphically intuitive way”.
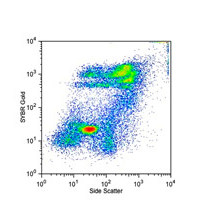 One of the most revolutionary developments in environmental sciences is the discovery of staggering molecular diversity among microbial populations. These organisms are primarily responsible for driving ecosystem structure and function, through the activation and regulation of a diverse array of molecular activities. Technological advances in analytical flow cytometry and cell sorting are rapidly revolutionizing our understanding of this genetic diversity and its relationship to function in widely varied environmental systems. Such analyses have provided unprecedented access to novel groups of organisms with unique molecular capabilities. Indeed, the recent discovery of the two most abundant phytoplankton in the world oceans (Prochlorococcus and Synechococcus) was only made possible by this analytical technique. The ability to now isolate specific subpopulations from natural communities and elucidate how they structure environmental niches and dictate response to environmental change represents a truly transformative time in microbial and molecular ecology.
The technological advances in flow cytometry and high-speed sorting and its recent in situ application to environmental microbiology and molecular ecology have made it an indispensable component for cutting edge research in these fields. Direct and convenient access to a flow cytometer/high-speed cell sorter, which is dedicated and calibrated for the analysis of environmental samples, is essential to elevate the quality of and provide new directions for ongoing research and graduate education in molecular ecology at Rutgers. Its creative implementation to novel research questions and its inclusion in graduate education and training is also essential to improve the quality of graduate programs, to recruit the highest caliber graduate students, postdoctoral researchers and faculty members, and to propel its research and education to the forefront of top tier programs.
One of the most revolutionary developments in environmental sciences is the discovery of staggering molecular diversity among microbial populations. These organisms are primarily responsible for driving ecosystem structure and function, through the activation and regulation of a diverse array of molecular activities. Technological advances in analytical flow cytometry and cell sorting are rapidly revolutionizing our understanding of this genetic diversity and its relationship to function in widely varied environmental systems. Such analyses have provided unprecedented access to novel groups of organisms with unique molecular capabilities. Indeed, the recent discovery of the two most abundant phytoplankton in the world oceans (Prochlorococcus and Synechococcus) was only made possible by this analytical technique. The ability to now isolate specific subpopulations from natural communities and elucidate how they structure environmental niches and dictate response to environmental change represents a truly transformative time in microbial and molecular ecology.
The technological advances in flow cytometry and high-speed sorting and its recent in situ application to environmental microbiology and molecular ecology have made it an indispensable component for cutting edge research in these fields. Direct and convenient access to a flow cytometer/high-speed cell sorter, which is dedicated and calibrated for the analysis of environmental samples, is essential to elevate the quality of and provide new directions for ongoing research and graduate education in molecular ecology at Rutgers. Its creative implementation to novel research questions and its inclusion in graduate education and training is also essential to improve the quality of graduate programs, to recruit the highest caliber graduate students, postdoctoral researchers and faculty members, and to propel its research and education to the forefront of top tier programs.
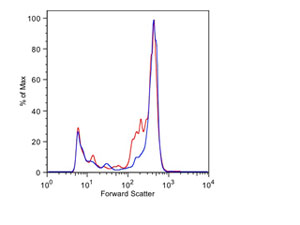 The unrivaled capabilities of the InFlux Model 209S Mariner as an extremely sensitive analyzer and an unparalleled cell sorter allows for the implementation of a wide array of multi-color, fluorescence-based analyses to environmental microbiology and molecular ecology. Its application involves the use of diagnostic, fluorescent markers (e.g., stains, gene expression constructs) in environmental microbes, including viruses, bacteria, fungi and phytoplankton, followed by detection and sorting for subsequent culturing, biochemical analysis, or gene expression studies. This is an extremely powerful approach for targeting and specifically interrogating the in situ abundance and activity of subpopulations of cultures or natural assemblages, and of assessing the importance of specific genes to environmental function. Moreover, the ability to sort and collect cells of interest with high purity opens up a vast array of post-sort possibilities, including proteomic and genomic analyses. Importantly, having the InFlux on site at IMCS, and within walking distance of other SEBS academic departments, provides the potential to analyze and sort live cells, absent of harsh fixatives and preservatives. Moreover, the custom microbiology and marine options on this InFlux instrument such as a small particle detector and a Lexan isolation hood and support frame for mounting on board a research vessel, provides heretofore-unobtainable capabilities for analyzing for analyzing and sorting natural populations in situ.
The unrivaled capabilities of the InFlux Model 209S Mariner as an extremely sensitive analyzer and an unparalleled cell sorter allows for the implementation of a wide array of multi-color, fluorescence-based analyses to environmental microbiology and molecular ecology. Its application involves the use of diagnostic, fluorescent markers (e.g., stains, gene expression constructs) in environmental microbes, including viruses, bacteria, fungi and phytoplankton, followed by detection and sorting for subsequent culturing, biochemical analysis, or gene expression studies. This is an extremely powerful approach for targeting and specifically interrogating the in situ abundance and activity of subpopulations of cultures or natural assemblages, and of assessing the importance of specific genes to environmental function. Moreover, the ability to sort and collect cells of interest with high purity opens up a vast array of post-sort possibilities, including proteomic and genomic analyses. Importantly, having the InFlux on site at IMCS, and within walking distance of other SEBS academic departments, provides the potential to analyze and sort live cells, absent of harsh fixatives and preservatives. Moreover, the custom microbiology and marine options on this InFlux instrument such as a small particle detector and a Lexan isolation hood and support frame for mounting on board a research vessel, provides heretofore-unobtainable capabilities for analyzing for analyzing and sorting natural populations in situ.
 One of the most revolutionary developments in environmental sciences is the discovery of staggering molecular diversity among microbial populations. These organisms are primarily responsible for driving ecosystem structure and function, through the activation and regulation of a diverse array of molecular activities. Technological advances in analytical flow cytometry and cell sorting are rapidly revolutionizing our understanding of this genetic diversity and its relationship to function in widely varied environmental systems. Such analyses have provided unprecedented access to novel groups of organisms with unique molecular capabilities. Indeed, the recent discovery of the two most abundant phytoplankton in the world oceans (Prochlorococcus and Synechococcus) was only made possible by this analytical technique. The ability to now isolate specific subpopulations from natural communities and elucidate how they structure environmental niches and dictate response to environmental change represents a truly transformative time in microbial and molecular ecology.
The technological advances in flow cytometry and high-speed sorting and its recent in situ application to environmental microbiology and molecular ecology have made it an indispensable component for cutting edge research in these fields. Direct and convenient access to a flow cytometer/high-speed cell sorter, which is dedicated and calibrated for the analysis of environmental samples, is essential to elevate the quality of and provide new directions for ongoing research and graduate education in molecular ecology at Rutgers. Its creative implementation to novel research questions and its inclusion in graduate education and training is also essential to improve the quality of graduate programs, to recruit the highest caliber graduate students, postdoctoral researchers and faculty members, and to propel its research and education to the forefront of top tier programs.
One of the most revolutionary developments in environmental sciences is the discovery of staggering molecular diversity among microbial populations. These organisms are primarily responsible for driving ecosystem structure and function, through the activation and regulation of a diverse array of molecular activities. Technological advances in analytical flow cytometry and cell sorting are rapidly revolutionizing our understanding of this genetic diversity and its relationship to function in widely varied environmental systems. Such analyses have provided unprecedented access to novel groups of organisms with unique molecular capabilities. Indeed, the recent discovery of the two most abundant phytoplankton in the world oceans (Prochlorococcus and Synechococcus) was only made possible by this analytical technique. The ability to now isolate specific subpopulations from natural communities and elucidate how they structure environmental niches and dictate response to environmental change represents a truly transformative time in microbial and molecular ecology.
The technological advances in flow cytometry and high-speed sorting and its recent in situ application to environmental microbiology and molecular ecology have made it an indispensable component for cutting edge research in these fields. Direct and convenient access to a flow cytometer/high-speed cell sorter, which is dedicated and calibrated for the analysis of environmental samples, is essential to elevate the quality of and provide new directions for ongoing research and graduate education in molecular ecology at Rutgers. Its creative implementation to novel research questions and its inclusion in graduate education and training is also essential to improve the quality of graduate programs, to recruit the highest caliber graduate students, postdoctoral researchers and faculty members, and to propel its research and education to the forefront of top tier programs.
 The unrivaled capabilities of the InFlux Model 209S Mariner as an extremely sensitive analyzer and an unparalleled cell sorter allows for the implementation of a wide array of multi-color, fluorescence-based analyses to environmental microbiology and molecular ecology. Its application involves the use of diagnostic, fluorescent markers (e.g., stains, gene expression constructs) in environmental microbes, including viruses, bacteria, fungi and phytoplankton, followed by detection and sorting for subsequent culturing, biochemical analysis, or gene expression studies. This is an extremely powerful approach for targeting and specifically interrogating the in situ abundance and activity of subpopulations of cultures or natural assemblages, and of assessing the importance of specific genes to environmental function. Moreover, the ability to sort and collect cells of interest with high purity opens up a vast array of post-sort possibilities, including proteomic and genomic analyses. Importantly, having the InFlux on site at IMCS, and within walking distance of other SEBS academic departments, provides the potential to analyze and sort live cells, absent of harsh fixatives and preservatives. Moreover, the custom microbiology and marine options on this InFlux instrument such as a small particle detector and a Lexan isolation hood and support frame for mounting on board a research vessel, provides heretofore-unobtainable capabilities for analyzing for analyzing and sorting natural populations in situ.
The unrivaled capabilities of the InFlux Model 209S Mariner as an extremely sensitive analyzer and an unparalleled cell sorter allows for the implementation of a wide array of multi-color, fluorescence-based analyses to environmental microbiology and molecular ecology. Its application involves the use of diagnostic, fluorescent markers (e.g., stains, gene expression constructs) in environmental microbes, including viruses, bacteria, fungi and phytoplankton, followed by detection and sorting for subsequent culturing, biochemical analysis, or gene expression studies. This is an extremely powerful approach for targeting and specifically interrogating the in situ abundance and activity of subpopulations of cultures or natural assemblages, and of assessing the importance of specific genes to environmental function. Moreover, the ability to sort and collect cells of interest with high purity opens up a vast array of post-sort possibilities, including proteomic and genomic analyses. Importantly, having the InFlux on site at IMCS, and within walking distance of other SEBS academic departments, provides the potential to analyze and sort live cells, absent of harsh fixatives and preservatives. Moreover, the custom microbiology and marine options on this InFlux instrument such as a small particle detector and a Lexan isolation hood and support frame for mounting on board a research vessel, provides heretofore-unobtainable capabilities for analyzing for analyzing and sorting natural populations in situ. 