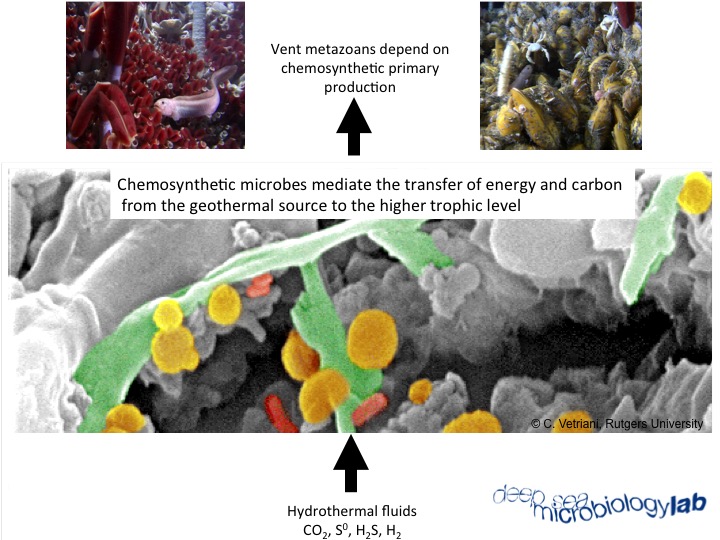
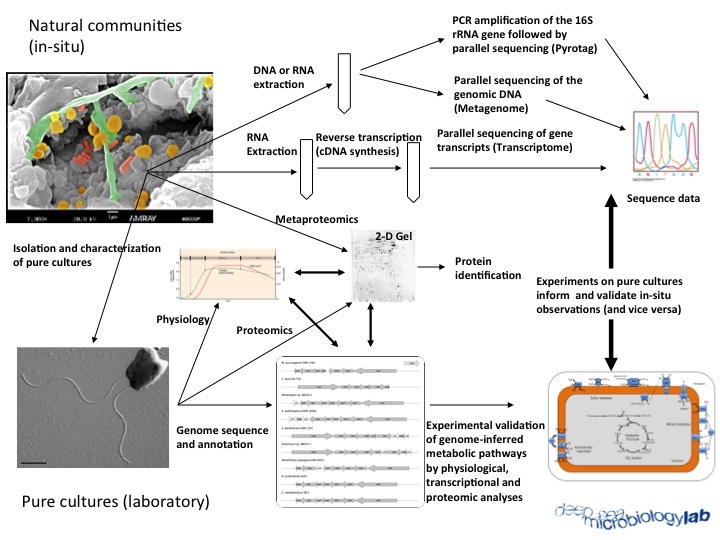
Integration of physiological and -omics studies to investigate natural microbial communities in marine geothermal environments
One approach in microbial ecology is to cultivate bacteria from their natural habitat and then determine their physiology and growth rates under conditions that simulate their natural environment. The key word here is “simulate”, because of the near impossibility to adequately replicate in situ conditions of nutrient flux, community structure, and environmental variables (e.g. temperature). A second approach is to use -omics techniques to investigate the distribution, community structure, metabolic potential and gene expression of microorganisms from environmental samples. At the DSML we integrate both approaches to investigate the ecology and function of natural microbial communities in marine geothermal environments.
Our investigation of nitrate reduction at deep-sea hydrothermal vents is one example of how we integrate physiology, genomic and metagenomic approaches. Work on pure cultures of vent bacteria and archaea indicated that nitrate respiration supports chemolithoautotrophic microbial growth in deep-sea geothermal environments. The analysis of the genome sequence of these organisms allowed us to reconstruct their nitrate reduction pathways and to identify key functional genes, e.g. the gene encoding the periplasmic nitrate reductase, napA. Subsquent culture-independent analyses of the napA gene revealed that nitrate respiration is conserved and widespread in vent Epsilonproteobacteria. For more information, read our paper in ISME J.
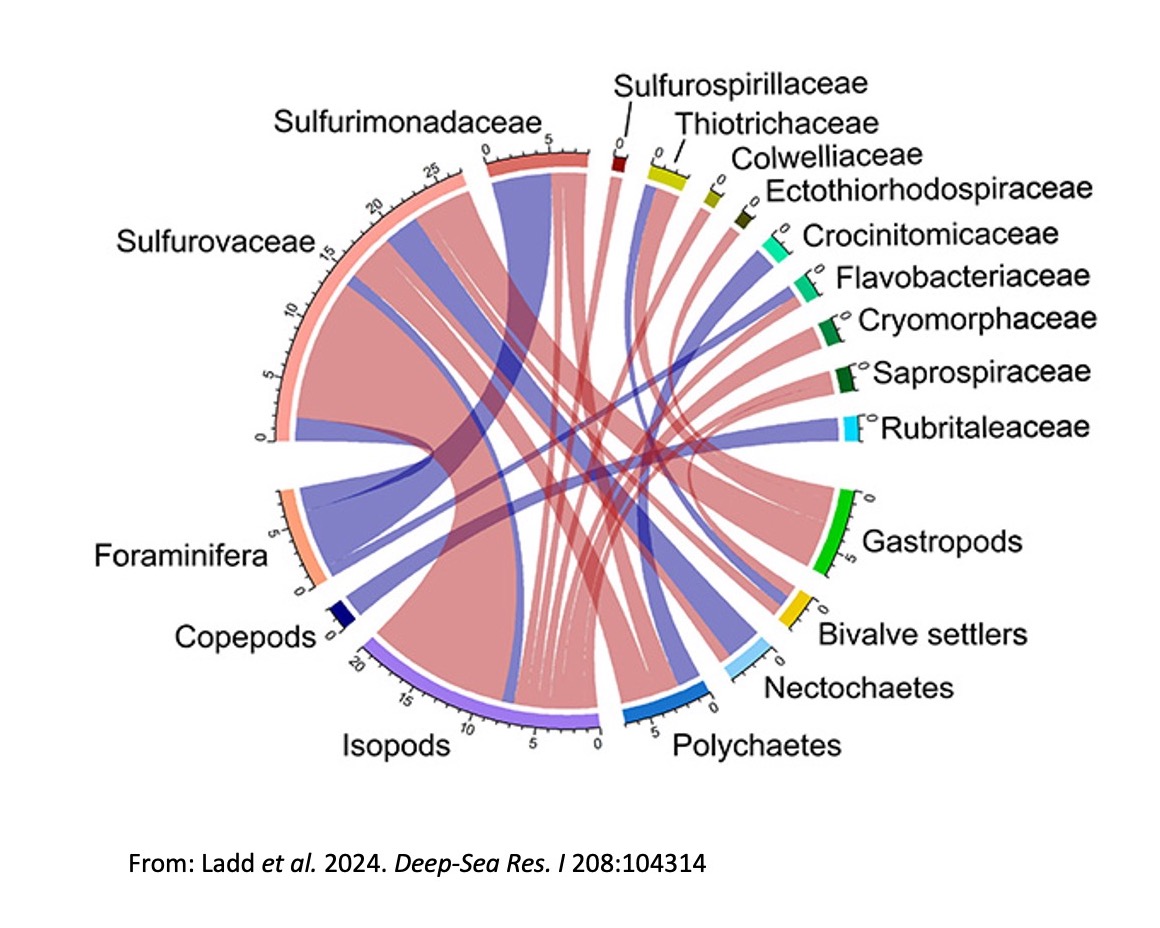
As part of our Biofilms4Larvae project, we investigated the role of chemosynthetic microbial biofilms as signposts for settlement of larvae at deep-sea hydrothermal vents. Our findings show that microbial community composition plays a role in larval settlement and animal migration in hydrothermal vent systems. The detection of microbial and faunal interactions provides a starting point for identifying key microbial characteristics influencing colonization processes at hydrothermal vents. For more information, read our papers in Deep-Sea Research, BMC Environmental Microbiome and Limnology and Oceanography Methods.